
Proteomics—the science of proteins—is opening up a new world of discovery and understanding of diseases as diverse as cancer and dementia. Using a variety of rapidly developing technologies, and knowledge gleaned, in part, from the successful effort to sequence the human genome, researchers the world over are developing new drugs and diagnostic tests based on proteins that are key to health as well as disease.
One of those methods, called “molecular fingerprinting,” attempts to identify patterns of proteins in the blood and tissues that can be used to detect diseases like cancer much earlier and monitor therapy much better than is now possible. This is the story of some of the research—and the essential involvement of a patient—that are helping to make that hope a reality.
Proteomics (pronounced “pro-tee-ohm-ics”) represents a growing understanding of the power of proteins to determine health and disease. These are not the proteins found in steak or peanut butter. They’re already in your body, making up the walls of your cells and tissues, transmitting electrical signals in your brain, and carrying out a host of other genetic instructions that are essential for life.
When they go wrong—perhaps because they’re mutated by a genetic error or overproduced—proteins can cause a wide range of problems, from heart attacks to Alzheimer’s disease.
Thanks to the technological revolutions of the past quarter century that enabled scientists to sequence the human genome and put vast amounts of computing power on a chip, the previously unseen world of proteins is being revealed. That knowledge is fanning the flames of a new revolution—one in which diseases may be diagnosed earlier, treated much more effectively and ultimately stopped in their tracks.
One of the first clinical applications of proteomics is the early diagnosis of cancer.
Scientists around the world are trying to define “molecular fingerprints” of various cancers—unique patterns of proteins that may signal the presence of tiny tumors not yet detectable by X-rays. The information could help doctors make an earlier diagnosis and determine in advance which treatment will be most effective. It also may lead to the development of new drugs that can—like precision bombs—knock out various steps in the cascade of events leading to malignancy, and without harming normal tissue.
Researchers don’t yet know the identity of all of the proteins. Nor do they understand what the spectrum of proteins suggests. But they’re learning, thanks to tissue donated by cancer patients. At Vanderbilt University Medical Center, for example, David Carbone, M.D., Ph.D., is leading a “molecular fingerprinting” study of lung tumors.
“We’re trying to determine if we can identify patterns at the time of diagnosis that can predict how a tumor’s going to behave,” says Carbone, Harold L. Moses Professor of Cancer Research. “If you knew this tumor was going to respond to chemotherapy, then you might consider giving adjuvant chemotherapy after surgery.”
Similar studies of cancerous breast, prostate and rectal tissue are being conducted at Vanderbilt. They join dozens of studies underway around the world. There is some early evidence that such studies can have powerful predictive value.
A complex world
Just what is this new science of proteins? How did we get here, and how far and how fast can we go?
The importance of proteins as the basic building blocks of life has been appreciated for more than 150 years (see “The Power of Proteins”), but until recently, the characterization of these fascinating molecules was a slow and arduous process. A scientist could spend a career trying to isolate, identify and understand a single protein—out of the hundreds of thousands that make up the human “proteome.”
Then came the fruits of the genomic and computer revolutions—methods for cloning genetic material (the “DNA”) and mass producing large quantities of it; automation and miniaturization; and the ability to create and sift through huge “libraries” of data on genes and proteins.
The recent sequencing of the 35,000 or so genes that make up the human genome has created a vast pool of information from which scientists hope to fish out ways to prevent dementia, cure cancer, and perhaps even eliminate ancient afflictions of the developing world.
For example:
- At the University of Virginia, Donald Hunt, Ph.D., and colleagues have identified peptides—fragments of proteins—that trigger the body’s immune system to kill melanoma (skin cancer) cells. This could lead to the development of an effective vaccine against the disease.
- Researchers at Stanford University have created a microarray or glass slide containing thousands of molecules that can bind to antibodies in the blood. It’s being tested as a way to improve the diagnosis of autoimmune diseases like rheumatoid arthritis, in which the body’s immune system attacks its own tissues.
- Scientists at Johns Hopkins University have developed a new blood test for malaria using mass spectrometry, a way of measuring and identifying proteins. The method could lead to improvements in early diagnosis and treatment of the disease, which kills more than a million people in equatorial countries every year.
- An “electronic taste chip” has been developed at the University of Texas at Austin that mimics the ability of the human taste bud to rapidly detect proteins and other chemicals in environmental samples. One possible application: detection of biological or chemical weapons.
As tantalizing as these examples are, few scientists predict that the discovery and understanding of important proteins will be as straightforward as the decade-long effort it took to read our genetic script. That’s because the DNA is more like a guidebook than a blueprint.
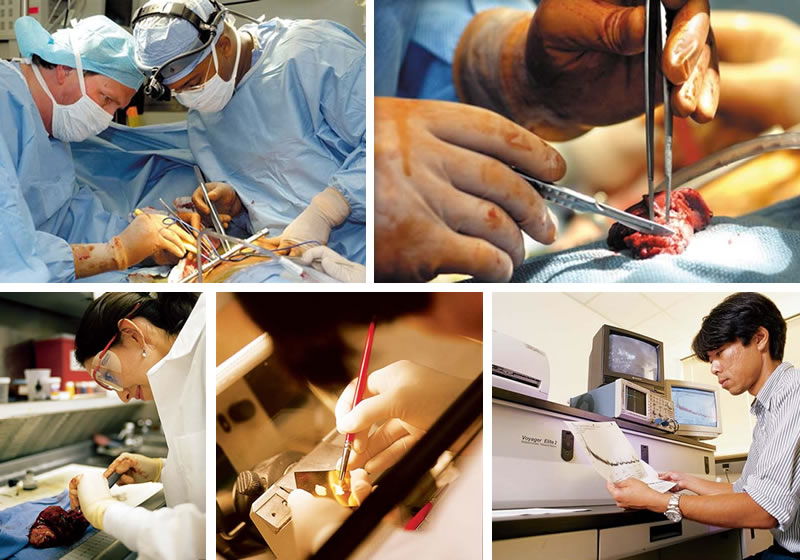
- John Roberts, M.D., left, chief of general thoracic surgery, removes a lung tumor with the help of surgery resident John Stewart, M.D.
- The tumor, removed from the left lung, is about an inch long.
- Adriana Gonzalez, M.D., examines a lung tumor in the surgical pathology lab.
- Slices of a tumor are prepared for a mass spectrometry study.
- Kiyoshi Yanagisawa, M.D., Ph.D., reads the mass spectrum of proteins in a tumor.
While humans apparently have fewer genes than it takes to make a rice plant, there are hundreds of thousands of different human proteins, perhaps more than a million.
The diversity and complexity of proteins is absolutely mind-boggling.
Insulin, which carries sugar into the cells for fuel, is made up of two polypeptide chains, one with 30 amino acids and the other with 21. Hemoglobin, the oxygen-carrying protein in red blood cells, is a complex three-dimensional molecule with four chains, each more than 140 amino acids long. The epidermal growth factor receptor, a target for some of the new cancer drugs, is a single-chain protein with nearly 1,200 amino acids (see “One Protein’s Story”).
Proteins also have many different jobs. They form the elastic and resilient framework of muscles, nerves and other body tissues. They carry signals within and between cells, and — in the case of antibodies — sound the alarm when a germ invades. Many of them are enzymes, catalyzing chemical reactions. Still others help “read” the genetic code so it can be “translated” into more proteins.
Scientists used to think that for every gene there was only one protein. They now know that while genes determine the sequence of amino acids that make up the protein backbone, the genetic instructions can be translated and implemented in more than one way.
Once produced, proteins can be modified—by the addition of a phosphate or sugar molecule, for example—in ways that change their shape and function. And while our double-helical string of genes is relatively static and unchanging, proteins are constantly in motion. Some proteins come on the scene just long enough to do their jobs, and then — in a blink of an eye — they’re gone.
Different parts of the body have different populations of proteins. They change, too, in response to environmental influences, like the digestion of a piece of cherry pie—or even the time of day. “Your proteins won’t be the same tonight as they are now,” says Richard Caprioli, Ph.D., director of the Mass Spectrometry Research Center at Vanderbilt.
Proteins aren’t lone wolves, either. They act and react as part of intricate networks and pathways that transmit signals to and from the DNA, across the cell membrane and through the blood to distant parts of the body. Thus, proteins have been called “molecular machines,” at work in cellular “factories.”
Just as proteins carry out many of the functions necessary for life, they also are at the root of many diseases. A form of diabetes, for example, results from an inadequate supply of insulin.
A change in a single amino acid in the oxygen-transporting hemoglobin molecule alters the protein’s three-dimensional shape, and results in sickle-cell anemia. Red blood cells with the abnormal protein are misshapen (sickle-shaped), break apart and block small blood vessels, causing pain and low blood count (anemia).
Too much protein also can be a problem. Elevated levels of the receptor for epidermal growth factor, for example, has been linked to a variety of cancers.
Molecular fingerprints
This connection between proteins and cancer, in particular, is driving a new growth industry in proteomics. Much of the effort is aimed at developing new drugs targeting specific proteins. But the search for new diagnostics is equally intense. That’s because some diseases, including some forms of cancer, typically escape notice until they are in an advanced, hard-to-treat stage.
Currently cancer is diagnosed through a variety of techniques, both non-invasive (ultrasound, X-rays, CAT scans, etc.) and invasive (primarily surgery). There are a few blood tests for cancer, which detect cancer-related antigens (proteins that bind with antibodies), but these are far from definitive.
For example, prostate cancer is associated with increased levels of prostate-specific antigen (PSA), but PSA levels also rise in response to exercise, infection and certain medications. Levels of cancer antigen 125, a screening tool for ovarian cancer, are abnormally high only about half the time in early disease.
Several years ago, researchers at the U.S. Food and Drug Administration and National Cancer Institute joined forces in an attempt to improve early detection of cancer, determine in advance which treatments are likely to be most successful in individual patients and, ultimately, to develop new “targeted” therapies that effectively stop cancer growth without harming normal tissue.
“Certainly cancer is underpinned by genomic disorders but functionally it’s a proteomic disease,” says Emanuel Petricoin, Ph.D., the project’s lead FDA researcher. “Effectively it’s the rewiring and miswiring of the protein circuits, the signal pathways, that cause cells to grow and not die.”
© 2001 Nature Publishing Group.
Lance Liotta, Ph.D., and his colleagues at the National Cancer Institute already had developed a microscope technique for teasing apart normal cells from their cancerous neighbors with the help of a laser. The cancer cells were then split open, and their contents poured onto a “protein chip,” a glass slide lined with “bait” molecules to which the cellular proteins stuck.
Extraneous material was washed away, and the slides were put into a mass spectrometer. As a first step, the slide was zapped with a laser, giving the proteins an electrical charge and spinning them off into a vacuum chamber toward an oppositely charged electrode. Their molecular weight was determined by the time it took them to get there.
The mass spectrometer then spit out a spectrum showing the molecular weights of all of the proteins and protein fragments. Dozens of experiments revealed that different cancer cells had unique spectra, or patterns of proteins, and thus potentially could be identified by their “molecular fingerprints.”
Using these techniques, the researchers are analyzing biopsies of cancerous tissue removed from patients before and after treatment to see if they can predict, from the protein patterns, which patients are likely to respond to specific drugs. The hope is that doctors one day will be able to “tailor” treatments to individual patients, while avoiding—in advance — therapies that won’t work.
The researchers also began testing the serum, the clear liquid part of the blood from cancer patients, to see if they could detect the characteristic protein patterns without actually removing any cancerous tissue.
In February 2002, they reported their first success. With the help of pattern-recognition computer software developed by scientists at Correlogic Systems in Bethesda, Md., the government researchers found unique patterns of five proteins in serum that could detect ovarian cancer with near-100 percent accuracy.
“Ovarian cancer is ripe for using this technique because it is usually diagnosed too late, when it’s almost untreatable, whereas if you catch it early enough, surgery alone can cure this disease most of the time,” Petricoin says.
Using a similar technique, researchers at Eastern Virginia Medical School in Norfolk have identified a pattern of nine serum proteins that accurately distinguish between prostate cancer, benign prostate hyperplasia (non-cancerous enlargement of the prostate gland), and healthy tissue.
The technique was considerably more specific than the PSA test, which has a high rate of false positive results, the researchers reported in 2002. If validated by further study, “this approach would have immediate and substantial benefit in reducing the number of unnecessary biopsies,” they concluded.
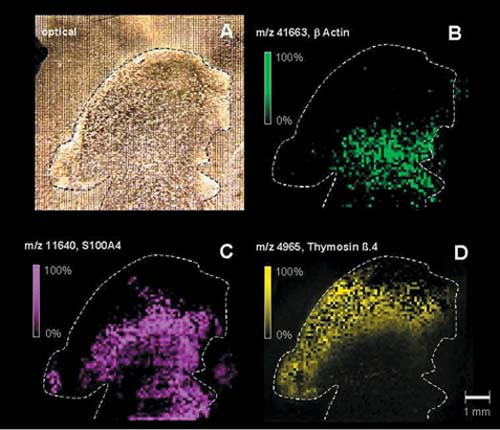
Caprioli and his colleagues have developed another mass spectrometry technique, called “imaging mass spectrometry,” that can actually “look” inside the tumor itself.
Using a laser and high-speed electronics and computers, they modified standard spectrometers so they could generate digital pictures, showing the distribution of individual proteins in cells and tissues. In April 2001 the researchers reported finding high levels of thymosin beta-4, a protein that may be a harbinger of malignant growth, in the outermost, proliferating edge of a human brain tumor that had been implanted in a mouse.
These “molecular photographs” may help improve the diagnosis and treatment of cancer. By pinpointing the precise location in the tissue where high levels of a protein are spurring cancerous growth, the technique could improve the accuracy of cancer surgery.
Repeat studies also can help determine how quickly tumors are growing, and whether they are responding to drug treatment, Caprioli says. This is “molecular insight that can be directly applied to patient care,” he says.
Petricoin acknowledges criticism that he and his colleagues have not identified the proteins they’re picking up in the blood. That will be crucial, other scientists say, for understanding the role the proteins may play in cancer growth, and how to develop new drugs to stop them. That’s true, agrees Petricoin, but patients need this technology now.
“We’re failing in the war against cancer,” he told a group of researchers at Vanderbilt in 2002. “For patients who have ovarian cancer … who have seen their mothers die of cancer, they don’t care what the underlying identity (of the protein) is. We have to test this hypothesis in the clinic. (But) we could use that pattern as a diagnostic today.”
Discovery science
“I don’t think they (the ‘omic’ strategies) will provide the answers to all of life’s mysteries,” responds Walter Chazin, Ph.D., Chancellor's Professor of Biochemistry and Physics and director of the Center for Structural Biology at Vanderbilt. “Nothing significant has been achieved yet … from using proteomics. It’s really in the technical development stage. The more we learn, the more complex it becomes.

Photo by Dean Dixon
“However, the one thing I do believe in is the value of these initiatives (to develop) high throughput technologies,” Chazin says. That’s the ability to accelerate the research process and generate huge amounts of data essentially overnight.”
The new technologies, in fact, are opening up a new way to conduct research (see “Discovery Science.”)
The traditional approach, exemplified by Stanley Cohen’s Nobel Prize-winning work at Vanderbilt four decades ago, is to begin with a biological observation. For example, Cohen noticed that an extract of mouse salivary glands causes newborn mice to open up their eyes earlier than normal.
That observation led to the discovery of epidermal growth factor (EGF) and, later, to the identification of the receptor through which it acts. Hypotheses were formulated to explain the phenomena that were observed, and experiments were designed to test them.
Meanwhile, certain cancers were found to have abnormally high levels of the EGF receptor. Technological advances enabled scientists to determine the three-dimensional structure of the protein, and to synthesize molecules that can block its activity. Cohen’s body of work, beginning with an incidental observation, helped lead to a new field of cancer research and drug development.
The tried-and-true hypothesis approach hasn’t been abandoned. But increasingly, scientists are first mining the genome and now the proteome for all the data they can. Rather than studying a car by kicking the tires, they’re taking the entire vehicle apart—including the engine—then fiddling with each part to see what it does.
“It’s like Magellan,” says Andrew Link, Ph.D., associate professor of Microbiology & Immunology and Biochemistry at Vanderbilt who has developed a new method for studying complex mixtures of proteins. “You send your boat out. You don’t know what you’re going to find.”
Pie in the sky

Photo by Dean Dixon
Disappointing results in recent clinical trials have tempered optimism about the ability of some of the early—and highly touted — “targeted” therapies to stop disease in its protein tracks.
In August 2002, the pharmaceutical giant AstraZeneca reported that adding Iressa, a drug that blocks the EGF receptor, to a standard cancer drug did not significantly improve survival in a large-scale study of patients with advanced non-small cell lung cancer, the most common form of the disease.
The two drugs may have antagonized each other’s effects, however. When given as a single agent, the response of tumors to Iressa “is still felt to be remarkable,” says Carbone, who directs a federally funded Specialized Program of Research Excellence in lung cancer at Vanderbilt. The drug, which has been given to more than 18,000 people worldwide, is at least two to three times better in treating recurrences of lung cancer “than anything else out there, as well as being convenient and safe to take,” he adds.
Simply blocking the EGF receptor may not be enough to stop the cascade of events that leads to cancer, however. “You give 100 patients Iressa, 15 percent will respond, and there’s no way for you to know right now which one,” Carbone says. “My belief is that cancer is more complicated than that … It might well be 50 genes that you’re going to have to look at simultaneously.”
The ultimate treatment may come in the form of “cocktails” of several drugs that target different genes and protein pathways, depending on the unique characteristics of the patient’s cancer.
“Different patients may respond to different cocktails, so the more you know about the biology of cancer, the more likely you are to be able to develop intelligent therapies,” Carbone says.
That’s why Carbone is carefully analyzing and cataloging the patterns of proteins found in the lung tumors removed from patients.
The patients will be followed and, over time, the researchers will see if there are unique patterns that correlate with specific outcomes, such as a recurrence of the cancer in the brain, response to chemotherapy, or poor survival. Eventually, they hope to be able to predict the course of a patient’s disease shortly after they remove his or her tumor, and plan treatment accordingly.
“If we know this tumor will have a propensity to go to the brain, we can give prophylactic cranial irradiation,” Carbone says. “If you knew this tumor was going to respond to chemotherapy, then you might consider giving adjuvant chemotherapy after surgery.”
Pierre Massion, M.D., assistant professor of Medicine and Cancer Biology at Vanderbilt, is working with Carbone on a related study—testing tissues removed from the lungs of smokers who do not have cancer, to see if they can predict which smokers are likely to develop cancer in the future.
“The whole paradigm has shifted,” says Carbone. “Ten years ago, we had a couple of drugs, they weren’t very effective and most people weren’t receiving them. Now we have a huge number of … interesting, molecularly targeted therapeutics out, and some of them are showing real clinical responses.
“I think a new era in lung cancer has started.”








