
The findings by Rachel Pharris, Chris Rife, and Richard Armstrong, Ph.D., defining the molecular structure of FosA appeared online in the Journal of the American Chemical Society on Aug. 20. (photo by Dana Johnson)
VUMC team probes bacteria secret sharing
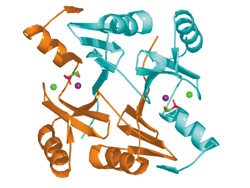
Ribbon diagram of the molecular structure of the FosA protein with fosfomycin (depicted in red and green) in the active site. The positions of manganese and potassium ions are shown as purple and green spheres, respectively.
Bacteria can do some pretty clever things to evade destruction by antibiotics, not the least of which is sharing biochemical secrets for elusiveness with other bacteria. It turns out that genetic messages can be passed between bacteria — even between different types of bacteria — that encode proteins enabling them to resist or destroy antibiotics.
In work that could lead to a better defense against just such a protein shared by a number of different disease-causing microorganisms, a group of Vanderbilt researchers has determined the crystal structure of a key antibiotic-resistance protein in Pseudomonas aeruginosa, a particularly dangerous and dreaded strain of bacteria.
The group, led by Richard N. Armstrong, Ph.D., professor of Biochemistry and Chemistry, has spent years studying this protein, called FosA, which confers resistance to the commonly prescribed antibiotic fosfomycin. Details of their work defining the molecular structure of FosA appeared online in the Journal of the American Chemical Society on Aug. 20.
Their intention, Armstrong said, is to make fosfomycin a more valuable drug by figuring out how to circumvent this resistance problem. Fosfomcyin is currently most useful as a treatment for urinary tract infections. Effective in a single dose usually given at a doctor’s office, it’s convenient and fast. But because the drug’s patent has expired, it’s not profitable. And because certain microorganisms that are common in urinary tract infections — E. coli, for example — may have the FosA protein, fosfomycin is not always effective.
In recent years, the DNA codes for a host of pathogenic microorganisms have been identified and posted in databases accessible to researchers worldwide. By searching these databases, Armstrong found the gene for FosA and similar resistance proteins in bacteria as diverse as Pseudomonas aeruginosa, Staphylococcus aureus, and Bacillus anthracis, the pathogen that causes anthrax.
Pseudomonas aeruginosa infects only immune-compromised individuals, such as patients with cancer or cystic fibrosis, and is notorious for being untreatable by any antibiotic. Staphyloccus aureus, a food-borne bacterium, is known to be resistant to a number of commonly used antibiotics, and that list is growing as the bacterium evolves.
“So there are a lot of high-profile microorganisms that have this resistance element in them,” said Armstrong. “And we believe, though we don’t have a large amount of evidence at this point, that these genes in the genomes of pathogens may confer intrinsic resistance to this particular antibiotic (fosfomycin), which renders it less effective.”
Chris Rife, a graduate student in Armstrong’s lab, set about determining how the FosA protein is built. The idea is, if you know what the building blocks are, how they fit together, and how the protein works to fend off antibiotic attack, you can design inhibitors of that ability. Rife was able to solve the unusual molecular structure of FosA by taking an original approach to a standard X-ray crystallographic method.
X-ray crystallography works by focusing X-rays at a crystalline protein sample and recording the diffraction pattern on a photographic imaging plate. Special computer programs interpret the resulting pattern of spots and convert it into a three-dimensional model. Of particular interest are the binding, or active, sites, the places in the protein molecule where interaction with other molecules makes different things happen.
From previous studies, the researchers knew that the FosA protein was a metalloenzyme, meaning it requires the presence of a metal — in this case, manganese — in its active site. For activation to occur, potassium must also bind at that site. In preparing his FosA crystals for study, Rife substituted the metal called thallium, which has binding qualities similar to potassium, and used a technique called multi-wavelength anomalous dispersion (MAD) to locate the potassium binding sites.
“MAD is a pretty common technique,” said Rife, “but no one had ever done it with this metal before. That made it kind of novel.”
Rife achieved his high-resolution characterization of the FosA protein by using a powerful, tunable X-ray source, called a synchrotron, located at the Argonne National Laboratory, a Department of Energy facility operated by the University of Chicago. A synchrotron is a huge ring, about three-quarters of a mile in diameter, with experiment stations spaced along the perimeter that tap off the X-ray beam line contained within it.
“The Argonne facility is great,” he said. “It’s an Advanced Proton Source that’s about seven years old, but it’s a half-million-dollar facility and the equipment is…just topnotch.”
The only drawback, said Rife, was having to work in a minimum of a 24-hour shift. To solve the structure of FosA, Rife drove several times to the Chicago lab — he couldn’t pass through airline security checkpoints with the tools he needed to take along — to work 48 hours straight, with no sleep, on each trip. “That wasn’t fun, but in the end it was worth it,” he said.
One of the surprising features of the FosA protein involves the manganese at its active site. Whereas most manganese-based metalloenzymes have six arms, or ligands, holding the metal in place, the manganese in FosA is anchored at only four points within the molecule.
“This is the first example of a manganese enzyme that has four rather than six coordinates,” said Armstrong. “I think this is chemically important because it makes the metal more reactive, so it facilitates the metal’s role in this (enzyme’s) reaction.”
The way that the FosA enzyme works is to bind the fosfomycin antibiotic at its active site and chemically destroy the antibiotic by adding a naturally occurring molecule called glutathione to it. The newly formed conjugate molecule lacks the power to disable the microbe.
Knowing the structure of the FosA active site and understanding the chemical action that occurs there hold the key to recovering the use of fosfoymycin as an effective antibiotic, and also open the possibility of designing new antibiotics.
“When an enzyme catalyzes a reaction, there’s a particular point — the transition state — that is the crucial structure…for the reaction to occur,” said Armstrong. “If we can figure out what the transition state looks like, we can tell (medicinal chemists) what kind of molecule they should build.”
That many microorganisms are resistant to fosfomycin may have an evolutionary explanation, one that involves that aforementioned ability of microbes to share genes. Long before fosfomycin was developed as an antibiotic for humans, it existed as a natural substance made by soil microbes to kill off other competing microbes.
“It’s part of the ancient arsenal of chemical weaponry that microbes use to fight each other for territory,” said Armstrong. “Because it’s a natural product, you’d expect that there might be a natural resistance that has evolved in various microorganisms.”
Bacteria are able to pass genes to one another by way of small circular pieces of DNA called plasmids. Armstrong believes that the FosA protein was likely shared among different human pathogens via plasmid vehicles. And, he says, that is maybe how resistance to antibiotics is spread among microorganisms in a hospital environment. “The hope is that finding inhibitors of resistance proteins will make existing antibiotics more effective.”
Other Vanderbilt researchers contributing to the published work include Rachel Pharris, a graduate student in the Chemical Biology graduate program who cloned and expressed the FosA gene from P. aeruginosa, and Marcia E. Newcomer, Ph.D., former professor of Biochemistry who is now a faculty member at Louisiana State University.
The work was supported by grants from the NIH and the U.S. Department of Energy.














