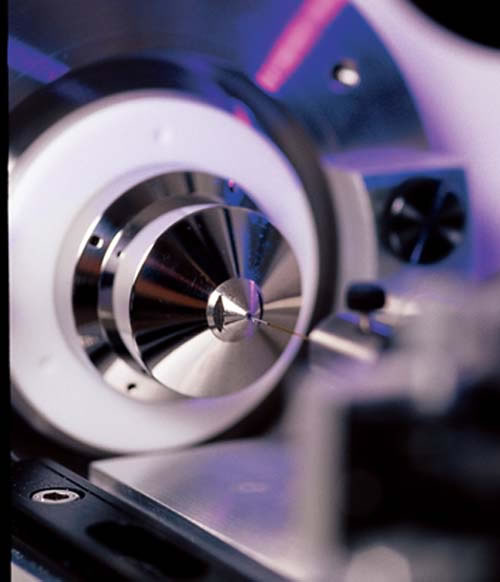
Proteomics is a technology-driven field. The photo at left, for example, shows proteins separated by high-performance liquid chromatography being sprayed into a tandem mass spectrometer, which will further separate them according to their “mass-tocharge” ratios. Here is a sampler of other important technologies:
Two-dimensional gel electrophoresis
This technique separates proteins across a gel, first according to their charge (isoelectric point) and then by their size (molecular weight). After separation the gels are stained, revealing dark spots where the proteins have concentrated. Spots of interest can be extracted from the gel and analyzed to identify the proteins.
In recent years several advances have improved the accuracy and sensitivity of 2-D gels. More reliable analytical methods, including robotic machines, have replaced hand-processed gels. By tagging proteins from two separate tissue samples with different fluorescent markers, researchers now can compare them on the same gel.
“This allows us to quantify the increase or decrease of a given protein,” in different stages of cancer, for example, says David Friedman, Ph.D., director of the proteomics laboratory in the Vanderbilt Mass Spectrometry Research Center. “It is very powerful at imaging thousands of proteins at a given time, yet it is biased toward more abundant proteins in the cell.”
Mass spectrometry
Mass spectrometry is becoming an increasingly valuable and versatile technique for identifying proteins, studying interactions between proteins, and mapping the location of proteins within tissue.
Ordinarily, proteins must be “cleaved,” or broken into manageable pieces called peptides, before they can be “weighed” in the mass spectrometer. The peptides are “ionized,” or given an electrical charge, so they can be propelled by an electric or magnetic field through a vacuum toward the instrument’s detector.
From the motion of the peptides, the spectrometer calculates their “mass-to-charge” ratio, which is related to their molecular weight, and generates a spectrum of the relative amounts (or intensity) of all the peptides that make up the original protein. The resulting peak-and-valley pattern of the mass spectrum is the protein’s unique “fingerprint.”
Techniques developed in the 1980s have greatly improved the ability of mass spectrometry to analyze large, intact proteins and complex protein mixtures.
Electrospray ionization, developed by John Fenn, Ph.D., and his colleagues at Yale University, is a method of squirting a protein solution through an intense electrical field in order to create a fine spray of charged droplets. The liquid evaporates, sending relatively large proteins into the mass spectrometer for analysis.
Another method, called matrix-assisted laser desorption/ionization or “MALDI,” was developed independently by researchers in Germany and Japan. Protein or tissue samples are mixed with a matrix, a material that can absorb laser energy, and applied to a target probe or plate. When struck by a laser, the matrix is partially vaporized, and electrically charged proteins are jettisoned into the mass spectrometer.
“It’s like hitting a golf ball out of a sand trap,” explains Vanderbilt mass spectrometry expert Richard Caprioli, Ph.D. “You hit the sand around it.”

For their contributions to mass spectrometry, Fenn and Japanese researcher Koichi Tanaka were awarded the Nobel Prize in Chemistry last fall. They shared the award with Kurt Wüthrich, Ph.D.
Detecting protein interactions
In the late 1980s, researchers at the State University of New York at Stony Brook pioneered a technique for studying protein interactions called the “yeast two-hybrid system.”
They broke apart a yeast protein that binds to and turns on a gene essential for cell growth. One half was attached to a known protein, or “bait,” and the other half was attached to the “prey,” the protein being studied. If the bait and prey interact, the two halves of the original protein will be brought back together, the gene will be turned on, and yeast cells will grow.
The technique does not pick up all of the interactions that occur in nature, but it has helped researchers construct “maps” of protein-protein interactions in organisms like the H. pylori bacterium, the major cause of ulcers. This information may lead to new ways to treat the infection.
Protein microarrays or “chips” provide another way to study interactions. The chips contain a variety of molecules that can bind specific proteins. Bound proteins, in turn, are detected through fluorescence labeling and other tagging techniques. This information is helping scientists understand how the cell operates at the molecular level, and how tiny changes in protein function can lead to disease.
Determining protein structure

X-ray crystallography uses X-rays to determine the three-dimensional structure of the crystallized form of proteins and other important biological molecules. The technique has led to the development of drugs that specifically bind to and inhibit disease-related enzymes, including one essential for production of the human immunodeficiency virus, which causes AIDS.
It can take days, weeks or even years to crystallize proteins in order to study them. “But the payoff for all of this effort is well worth it,” says Walter Chazin, Ph.D., director of Vanderbilt’s Center for Structural Biology. “X-ray crystallography, as the core technique of structural biology and structural genomics, is providing a paradigm shift in the ability of scientists to understand, control and design new biomedical activities.”
Another way to determine protein structure is to analyze the way the nuclei of the atoms that make up the molecule “resonate” under the influence of radio waves in ultra-high magnetic fields, a technique called nuclear magnetic resonance spectroscopy.
In the 1980s, Wüthrich worked out a method for interpreting the NMR spectra of proteins in solution. Since then, he and his colleagues have determined the NMR structure of more than 50 nucleic acids and proteins, including prion proteins, which are thought to cause mad cow disease.
Bioinformatics
Bioinformatics is the collection, organization and analysis of large amounts of biological information using networks of computers and databases. Vast, on-line “libraries” of confirmed protein sequences and the mass spectra “fingerprints” for thousands of proteins, predicted from the genes that encode them, have greatly improved the ability of researchers to identify proteins.
For example, researchers at the University of Washington in Seattle developed a computer program, called SEQUEST that helps researchers search databases rapidly for a possible “match” to the protein they’ve discovered.
Andrew Link, Ph.D., associate professor of Microbiology & Immunology and Biochemistry at Vanderbilt, uses SEQUEST in a technique he developed, called “direct analysis of large protein complexes” (DALPC).
Proteins are fragmented and the resulting peptides are separated by multi-dimensional liquid chromatography. This is a method for separating tiny amounts of peptides according to a variety of physical attributes, such as size or charge, as they travel through flexible plastic columns no bigger than the size of a human hair.
The peptides are analyzed by mass spectrometry, then fragmented to even smaller pieces and analyzed again to get information about their amino-acid sequences (a technique called “tandem” mass spectrometry). SEQUEST is then used to help identify them, thereby avoiding the need to purify each protein individually. DALPC “gives us the ability to look at complexes of hundreds of proteins at a time, and figure out what they are in a single experiment,” Link says.














